How Well Do Bio-Markers Work for CNN Training: A Comparison of Model Results
By Jacob Ryan
View Project GitHub
View Report
What is this?
This website aims to explain the project of its namesake. It involves deep learning (a form of machine learning commonly referred to as Artificial Intelligence) for the purpose of predicting whether chest radiographs (X-Rays) contain pulmonary edema. This site expects readers to have a basic familiriaty of what deep learning is and will explain the work with this idea in mind. However, any confusing terminology of this site can quickly be explained by a quick google search.
Why Deep Learning for Medical Imaging?
Medical imaging is costly and can take a lot of time to get results, as a specialist must analyze each image to determine what, if anything, is present. Because of this, there has been large strides to bring deep learning to medical imaging. Massive amounts of progress has been made, and there are many models that have fantastic levels of accuracy for determining what is present in a medical image.
Unfortunately, in order to train these models in the first place, researchers need access to thousand if not millions of high resolution medical images with their proper specialist given labels. The process of collecting a dataset like that is, of course, costly and time consuming (a cyclical issue).
An Brief Explanation of the Paper “Deep Learning Radiographic Assessment of Pulmonary Edema: Optimizing Clinical Performance, Training With Serum Biomarkers”
This paper was written by members of Augmented imaging/Artificial intelligence Data Analytics (AiDA) Lab, headed by Dr. Albert Hsiao. The findings are significant: models trained on chest radiographs to predict NT-proBNP (BNPP), a protein found in a blood serum test, levels performed very well. What does this mean? Well, BNPP levels are highly correlated with pulmonary edema, so much so that the levels of the protein found in the blood test are a biomarker of the condition. So, just by looking at XRays, where the pulmonary edema is either present or not, models were able to effectively predict BNPP levels. This then begs the question “Can models trained to infer BNPP levels accurately predict the presence of pulmonary edema?”
reexplain this section starting so just by looking at
Why Does It Matter If This Model Works?
If a model trained solely to infer the presence of pulmonary edema from chest XRays by utilizing a threshold of BNPP value and could generalize to perform well on radiologist confirmed pulmonary edema XRays, there would be less of a need to have radiologist labeled XRays in the first place. This utilization of biomarkers for the training process here (if successful) could set the stage for other medical imaging models to do the same. Moving towards biomarkers for ground truth levels would decrease the cost and time of acquiring large enough datasets for future experiments.
The Experiments
The Data
UCSD Data
‘UCSD’ Data is set of about 16,000 chest radiographs. Rather than having radiologist confirmed for whether the XRay contains pulmonary edema, the dataset (from AiDA Lab’s paper) contains only BNPP values. If the BNPP values is over a threshold of 400 pg/mL, the XRay was labeled as if it did contain pulmonary edema. Otherwise, it was labeled as if it did not.
UCSD Data Statistics
The threshold of 400 pg/mL was recommended by AiDA labs. This data had 64.6% positive labels across the set. The dataset is then split into train and validation sets with splits equal to 80% and 20%, respectively.MIMIC Data
‘MIMIC’ Data comes from MIT’s MIMIC-CXR public dataset, a project to provide anonymized chest radiographs and their respective medical results to the deep learning/medical imaging community. From this set, about 22,000 images were utilized. A portion of the MIMIC set is witheld and used as the test set all models were evaluated on.
MIMIC Data Statistics
MIMIC data had 48.7% positive labels across the set. The dataset is then split into train, validation, and test sets with splits equal to 80%, 10%, and 10%, respectively.Anatomical Segmentation
There’s an entire class of deep learning models called segmentation models. They are used to segment out a portion of the image. These are already incredibly popular; Apple photos uses it as part of the image cutout feature.

Using a segmentated image can help focus a classification model on the important aspects of the image by reducing the noise the model has to look at. Fortunately, AiDA Labs provided their anataomical segmentation model. All images are run through this model, providing an individual image of just the heart and an individual image of just the lungs. Using these focused images could improve model proficiency.
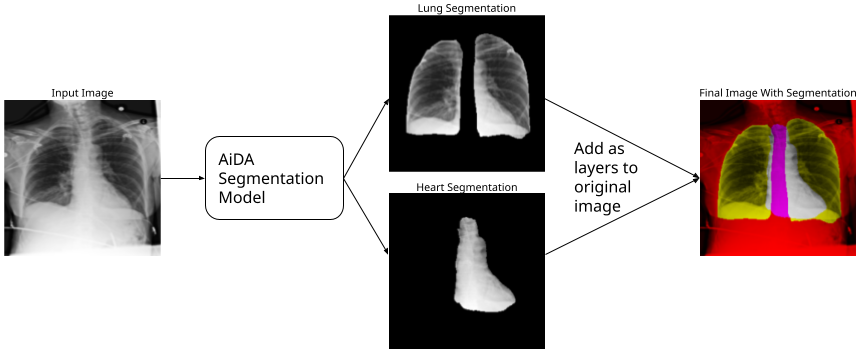
The Hypotheses
-
Deep learning models trained to infer the presence of pulmonary edema from chest XRays and BNPP levels (UCSD Data) rather than radiologist diagnoses will generalize to unseen data with radiologist diagnoses provided by MIMIC-CXR.
-
A deep learning model that emphasizes anatomic structures will perform better than one that is not informed. This will be evaluated by training two models for each dataset, two provided and tested on lung-and-heart segmentations as additional input channels and two without.
The Models
There are four models:
- UCSD Full: a model trained with full images and biomarker-based labels
- MIMIC Full: a model trained with full images and radiologist confirmed labels
- UCSD Segmented: a model trained with anatomically segmented images and biomarker-based labels
- MIMIC Segmented: a model trained with anatomically segmented images and radiologist confirmed labels
Each model is based on a ResNet 152v2 with pretrained weights from the ImageNet dataset.

What is ResNet 152v2 and what is ImageNet?
The ResNet 152v2 is an image classification model built by Microsoft that is renowned for its use of residual connections, a technique that allows networks to have a large amount of layers without losing its ability to predict. [ImageNet](https://www.image-net.org/about.php) is an industry standard dataset often used to benchmark classification model performance. Models trained to perform on ImageNet have developed a feature space that is adept at predicting many different classes of things that may be in the image. It is a great 'starting place' for models trained on niche tasks, like this one.
The Training Process
All models were trained for at max 35 epochs using a static learning rate of 1e
Biomarkers Did Not Work As Well, But Segmentation Might Help
There were three statistics used to represent the efficacy of the models: Area Under the Receiver Operator Curve (AUROC), Precision Recall Score (PRC), and Accuracy. All of these scores are a proportion, with ‘perfect’ performance being a 1.0 and the worst performance being a 0. AUROC represents the tradeoff between more correct postive predictions (true positives) and accidentally allowing more incorrect positive predictions (false positives). PRC represents the tradeoff between the proportion of true positives out of all positive guesses and the proportion of true positives out of all real positive labels. Accuracy, of course, is the proportion of labels that are correct.
| UCSD-Trained Full | MIMIC-Trained Full | UCSD-Trained Segmented | MIMIC-Trained Segmented | |
|---|---|---|---|---|
| AUROC | 0.755 | 0.877 | 0.754 | 0.889 |
| PRC | 0.590 | 0.799 | 0.559 | 0.816 |
| Accuracy | 0.652 | 0.793 | 0.616 | 0.802 |
With or without anatomical segmentation, MIMIC trained models outperformed UCSD trained models. Biomarkers, in this experiment, are not a sufficient replacement for radiologist labels for models to identify the presence of pulmonary edema. The figures below showcase this point.
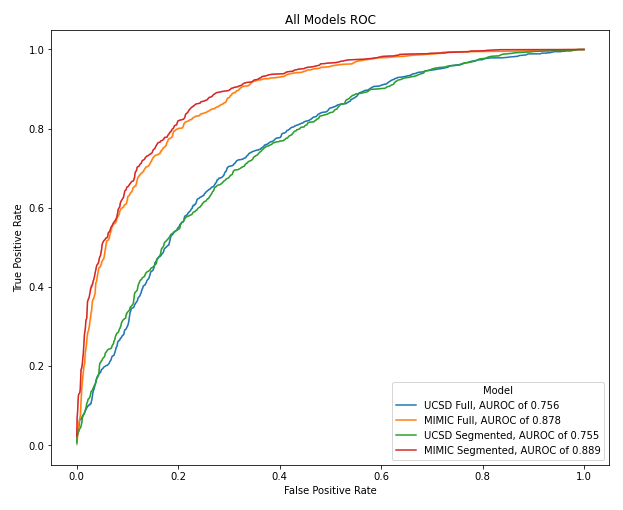
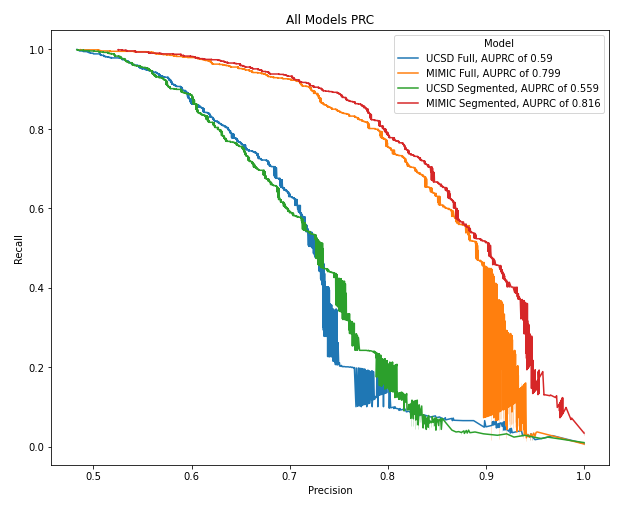
MIMIC trained models had atleast a 0.1 higher AUROC and PRC than UCSD trained models. Therefore, this experiment does not lead to the results that radiologist labels can potentially be phased out in favor of cheaper and fast biomarker levels.
As for anatomical segmentation, the results are mixed. The best overall model was the MIMIC Segmentation model, that is the model that was trained on anatomical segmentation images from the radiologist labeled dataset. This could lead to the conclusion that anatomical segmentation improves model efficacy. However, the UCSD Segmentation model underperformed its non-anatomical-segmentation counterpart. The results are gray and require further testing before anything is ruled out.
Potential Problems with this Experiment
There are a few reasons why MIMIC trained models outperformed UCSD models. First, biomarkers are a good correlative feature for pulmonary edema identification, but with enough error to affect model performance significantly. In this case, radioligist confirmed ground truths are to be preferred. Second, models trained on biomarkers need additional hyper-parameter-optimization to perform at the level of models trained on radiologist confirmed ground truths. This experiment trained all models using the same techniques for fairness; however, this could have the exact opposite effect by not giving enough accommodation to a model that is performing a harder task.
The best performing model, the MIMIC Segmentation model, was further analyzed using saliency techniques.
Saliency
Saliency techniques are a way of visually interpreting what the models are looking to for their predictions. Deep learning models inherently translate human problems (identification of pulmonary edema) into a new dimensional space only computers can understand. Saliency techniques help translate the computer understand back for human analysis. These techniques can serve as gut checks that the model is looking at appropriate regions, help analyze where model predictions go wrong, and see what artifacts from the image the model picked up on that humans could not.
This project utilized the XRAI (not XRay) technique.
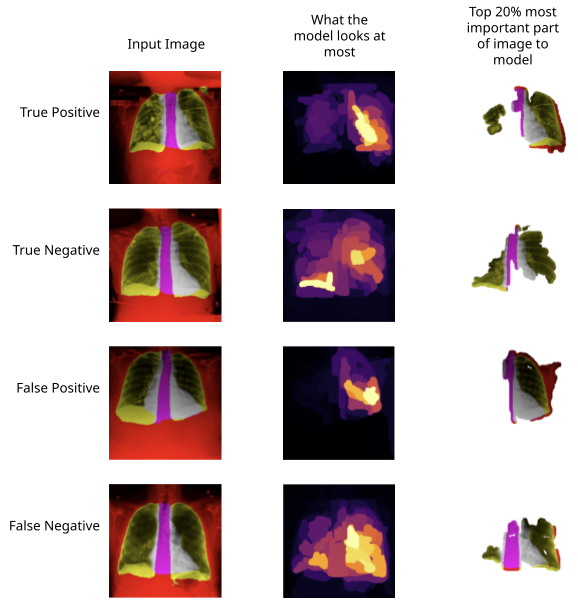
Here we can see what the model looks to when it gets a positive prediction right, a negative prediction right, a positive prediction wrong, and a negative prediction wrong. For positive predictions, the model seems to focus on a lung as a whole along with the majority of the heart. Negative predictions tend to look at the overlap of heart and left lung rather than the whole lung.